| Technical Name | High-through-put drug development of SARS-CoV-2 infection | ||
|---|---|---|---|
| Project Operator | National Yang-Ming University | ||
| Project Host | 黃琤 | ||
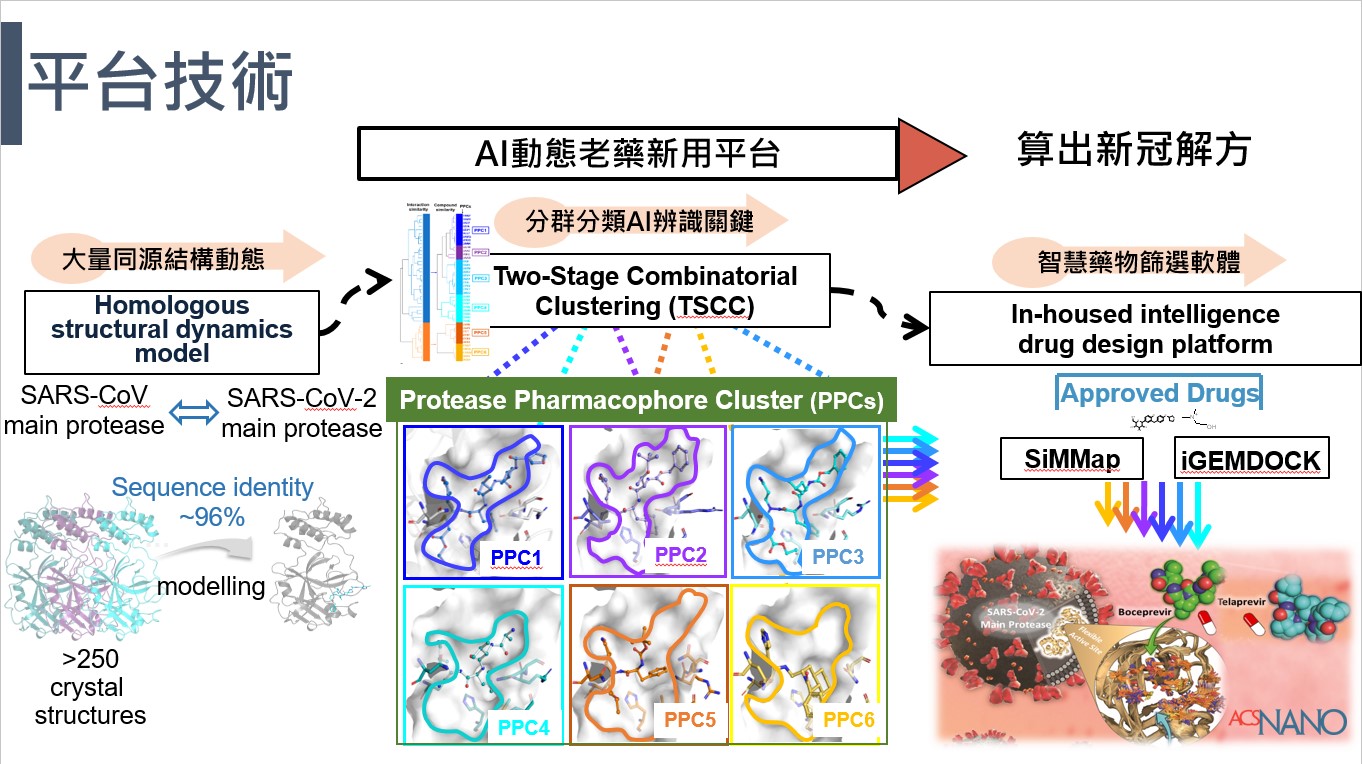
| Summary | Acknowledging the structural domain, the 3CLpro protein sequence of SARS-CoV was compared with that of SARS-CoV-2, yielding a sequence identity of 96% where alterations were found at T35V, A46S, S75N, L86V, R88K, S94A, H134F, K180N, L202V, A267S, T285A and I286L 6. Chang, et al. 7 had ranked the affinity of 10 FDA-approved HIV protease inhibitors to the 3CLpro in SARS-CoV-2 using computer-generated molecular docking and structural characteristics, while Nguyen, et al. 8 screened 1465 FDA-approved drugs in silico and listed the top 20 candidates that could inhibit 3CLpro of SARS-CoV-2. |
||
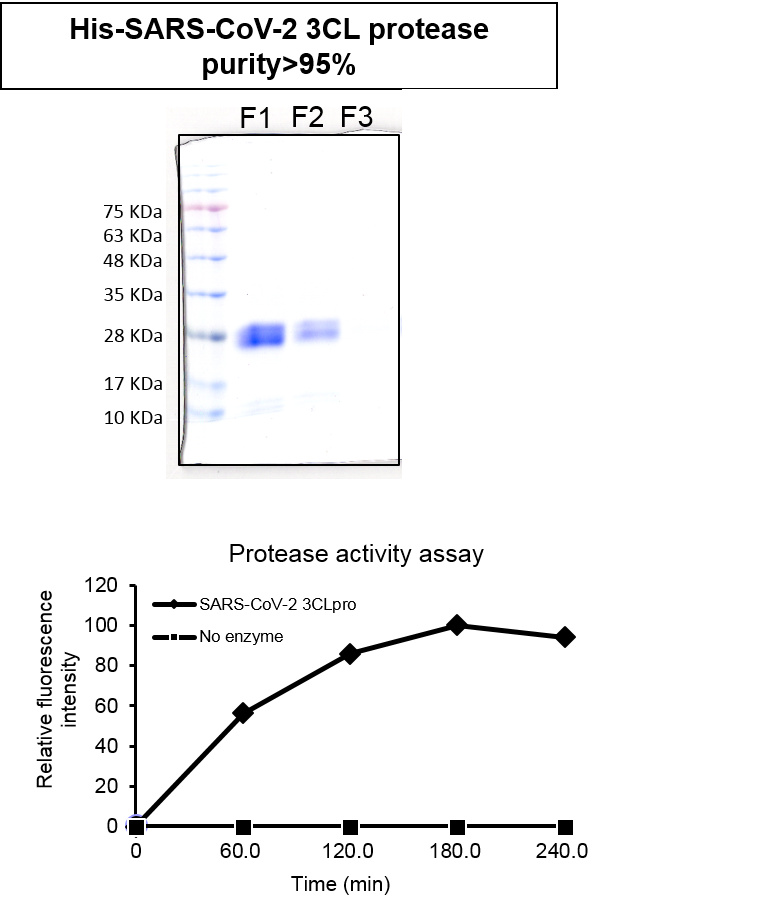
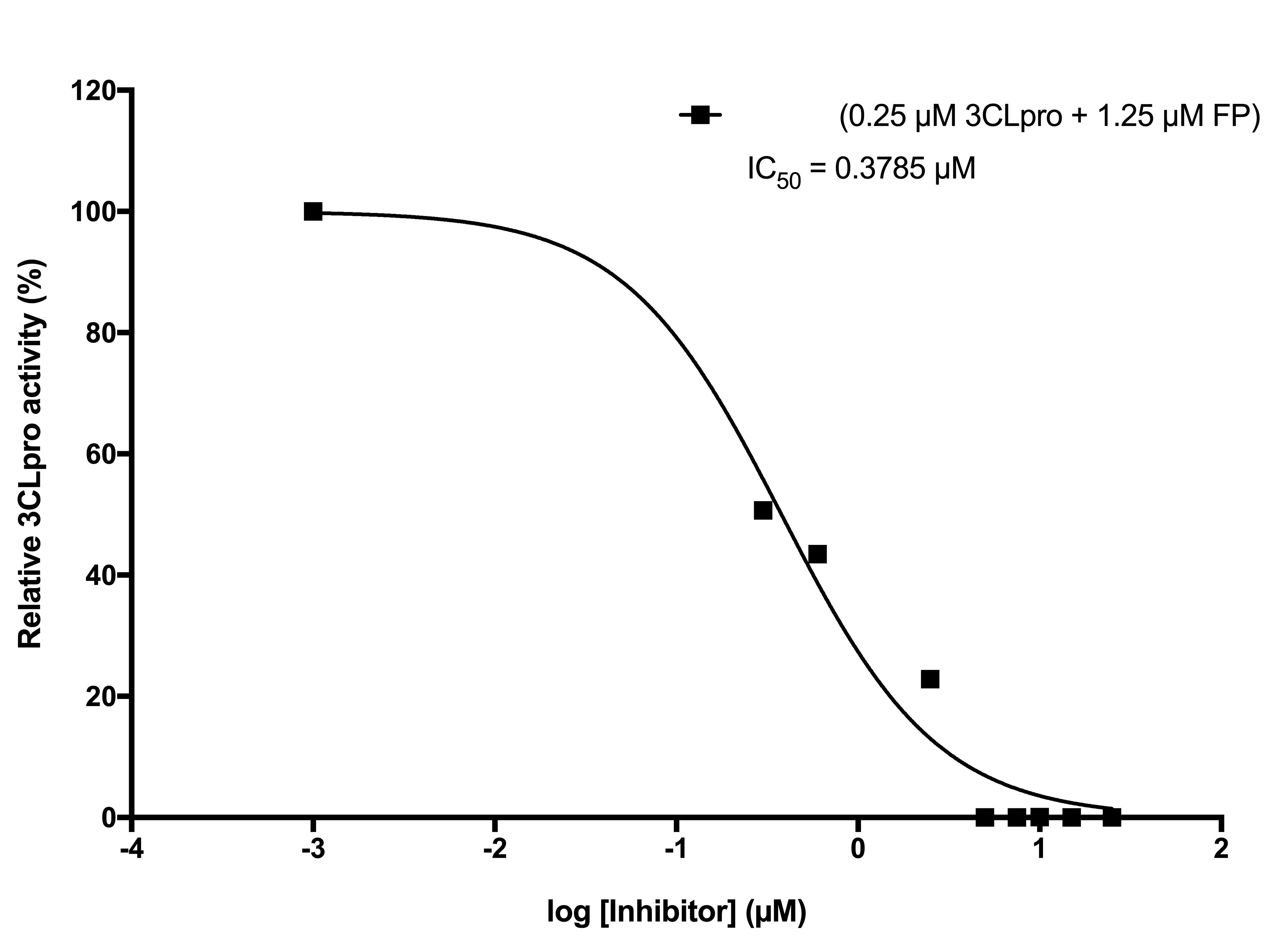
| Scientific Breakthrough | Considering that the outcomes of the clinical trials of lopinavir and ritonavir were negative in treating COVID-19 patients 9 and that remdesivir is still under clinical evaluation, the main objective of this study is to identify any FDA-approved drugs, developed drugs, and natural compounds that can be agilely repurposed to COVID-19 treatment. The half maximal inhibitory concentration of compounds (IC50) that inhibit SARS-CoV-2 3CLpro activity will also be determined. Concerning the urgency and the desperate needs for effective therapeutics, a high-throughput drug screening platform is carried out by using intramolecular quenching of fluorescence (IQF)-based fluorescence resonance energy transfer pair (FRET) technology. |
||
| Industrial Applicability | The half maximal inhibitory concentration of compounds (IC50) that inhibit SARS-CoV-2 3CLpro activity will also be determined. Concerning the urgency and the desperate needs for effective therapeutics, a high-throughput drug screening platform is carried out by using intramolecular quenching of fluorescence (IQF)-based fluorescence resonance energy transfer pair (FRET) technology, in which the quenched fluorescence is released upon 3CLpro-mediated proteolysis. That is, agents that to some extent induce inhibition of SARS-CoV-2 3CLpro lead to a decrease in the fluorescence intensity, as opposed to drugs that are of no avail. |
||
| Keyword | SARS-CoV-2, COVID-19 treatment, repurposing drugs, Drug development | ||
- chengh@ym.edu.tw
other people also saw