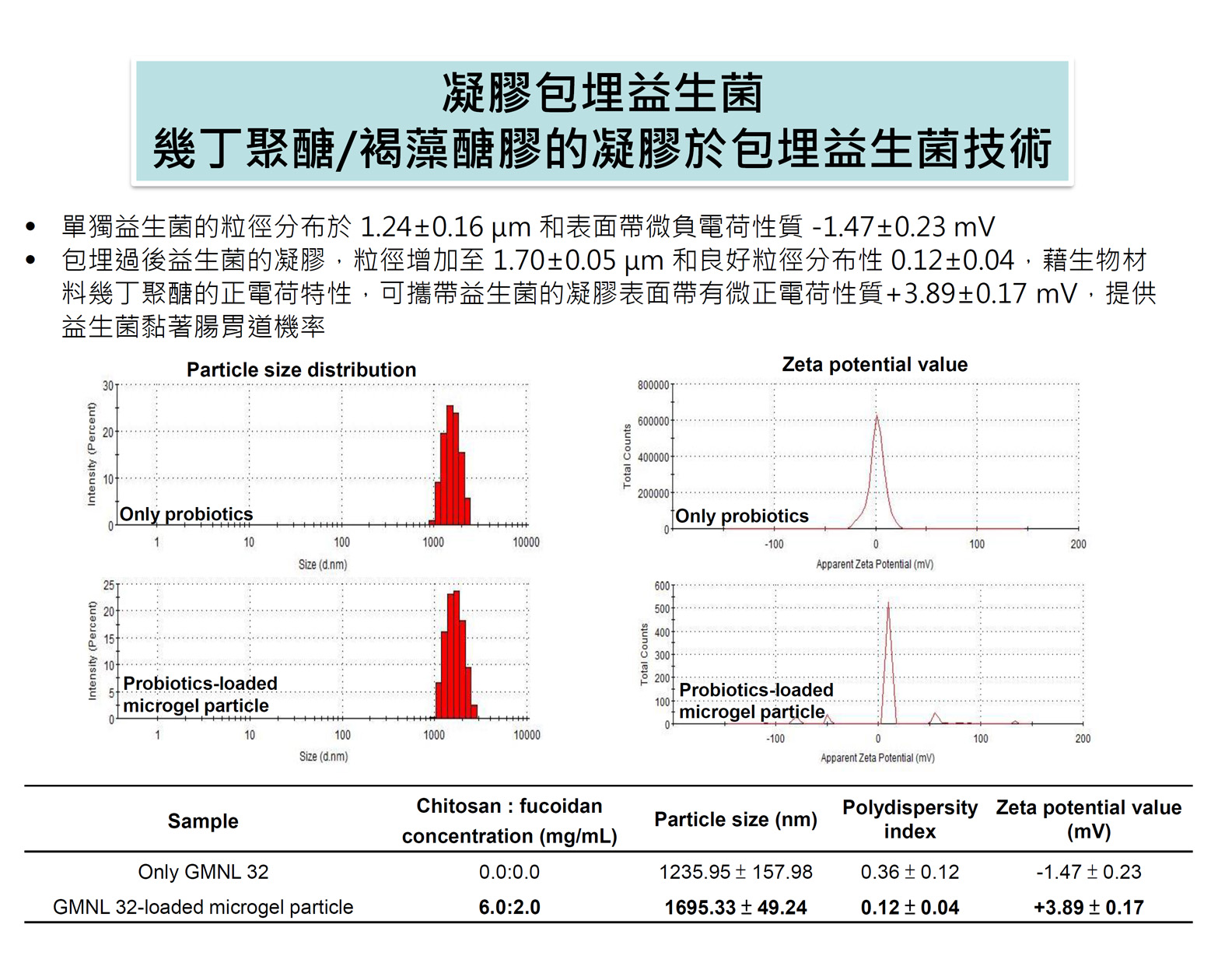
| Technical Name | Determination of genotypic resistance of Helicobacter pylori | ||
|---|---|---|---|
| Project Operator | National Taiwan University Centers of GenomicPrecision Medicine | ||
| Project Host | 陳培哲 | ||
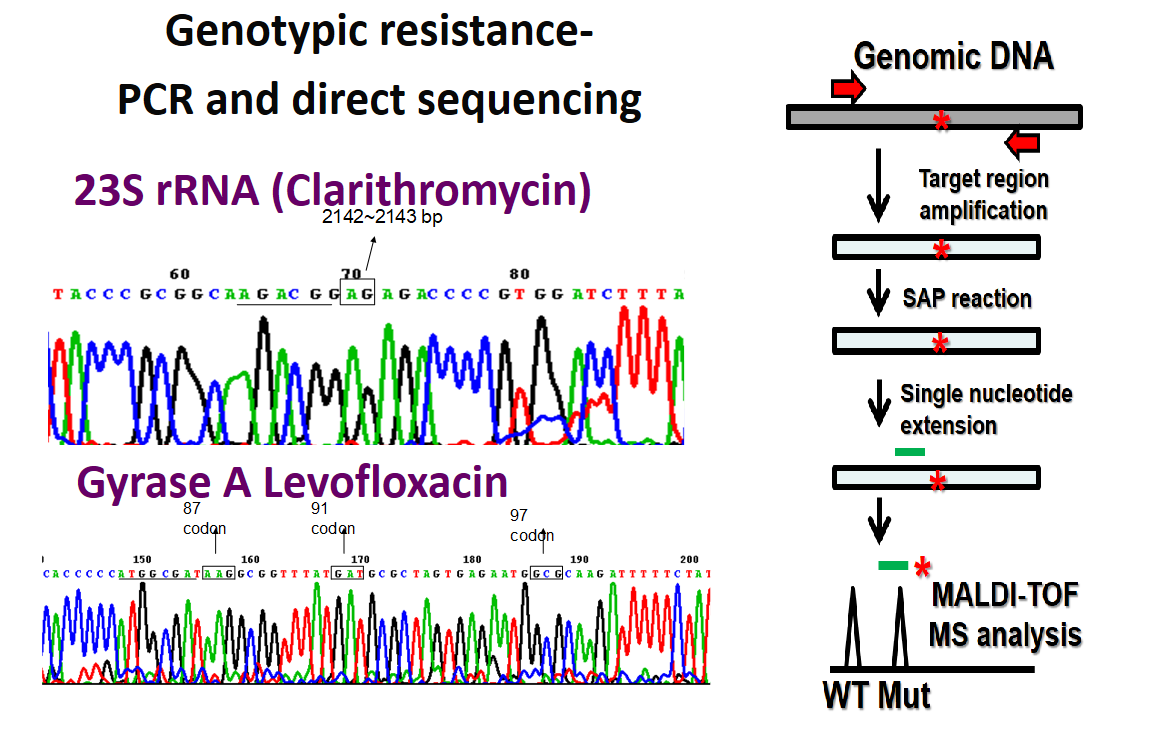
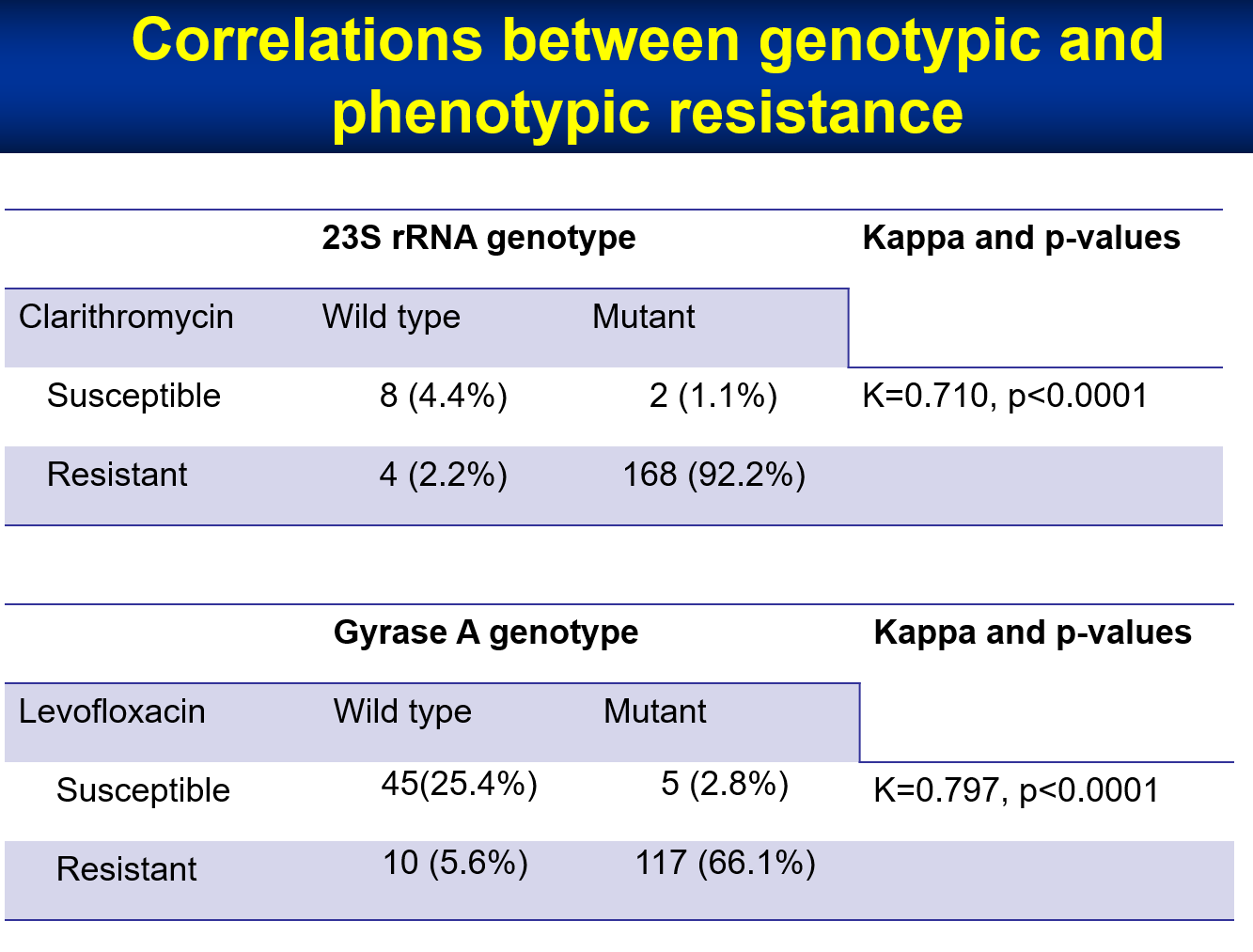
| Summary | We determined the point mutations of 23S rRNAgyrase A. We showed that the point mutations at 21422143 of 23S rRNA correlated well with clarithromycin resistance of H. pylori. Point mutations at gyrase A correlated well with levofloxacin resistance. The successful rate of genotypic resistance determination using gastric biopsy specimen was 98. |
||
| Scientific Breakthrough | Determination of the minimum inhibition concentration of antibiotics against H. pylori is time consumingthe successful culture rate is lower than 90. We showed that the point mutations at 21422143 of 23S rRNA correlated well with clarithromycin resistance of H. pylori. Point mutations at gyrase A correlated well with levofloxacin resistance. The successful rate of genotypic resistance determination using gastric biopsy specimen was 98. |
||
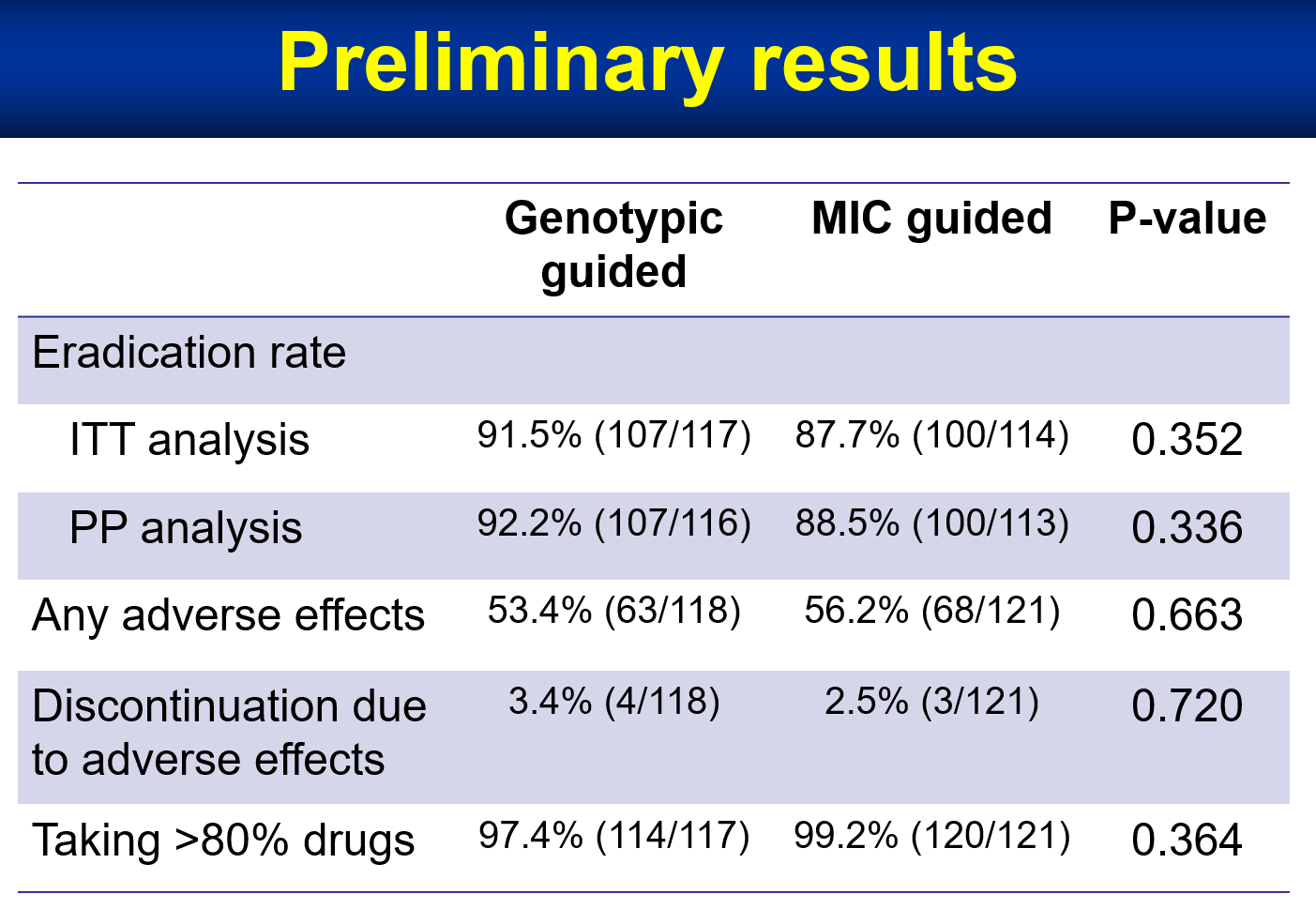
| Industrial Applicability | This technique can be used for tailored therapy for H. pylori eradication. We are developing techniques to determine the genotypic resistance of H. pylori using fecal samples. We expect that genotypic resistance guided therapy can improve the efficacy of H. pylori eradication therapyavoid the use of unnecessary antibiotics in clinical practice. |
||
| Keyword | H. pylori tailored therapy antibiotic resistance genotypic phenotypic 23S rRNA Gyrase A | ||
- Contact
- Yee-Shin Lee
- yslee@ntu.edu.tw
other people also saw