| Technical Name | Listening to the sea of biological big data (Seashell): an automatic web portal system for single-cell RNA sequencing (scRNA-seq) analysis | ||
|---|---|---|---|
| Project Operator | Academia Sinica | ||
| Project Host | 林仲彥 | ||
| Summary | Single-cell RNA sequencing (scRNA-seq) is now widely applied to biomedical researches. However, it is also difficult to handle the massive single-cell omics data without basic knowledge. To solve this problem, our research group developed an automatic web portal system for single-cell RNA sequencing analysis. |
||
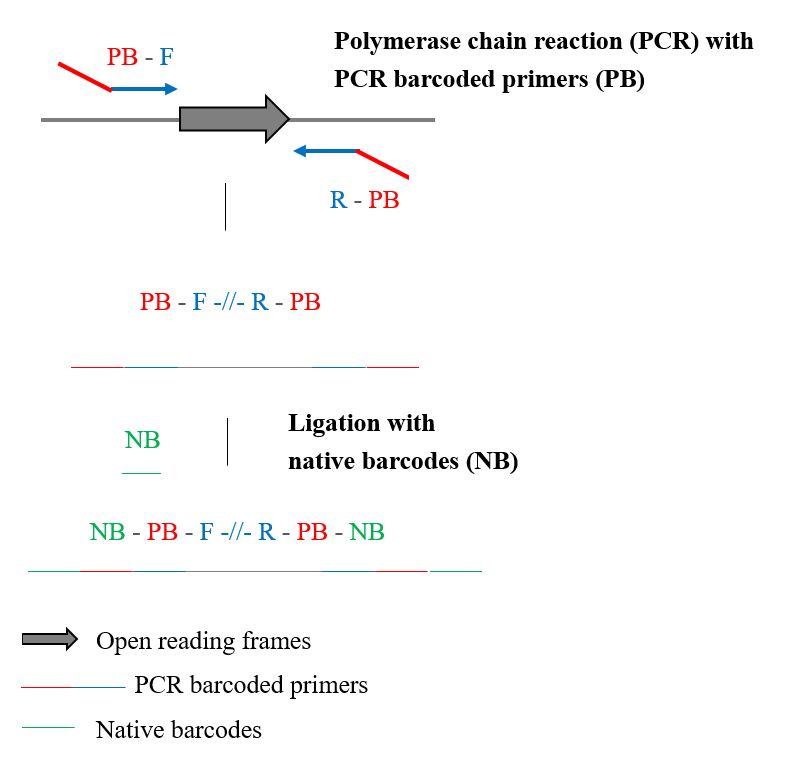
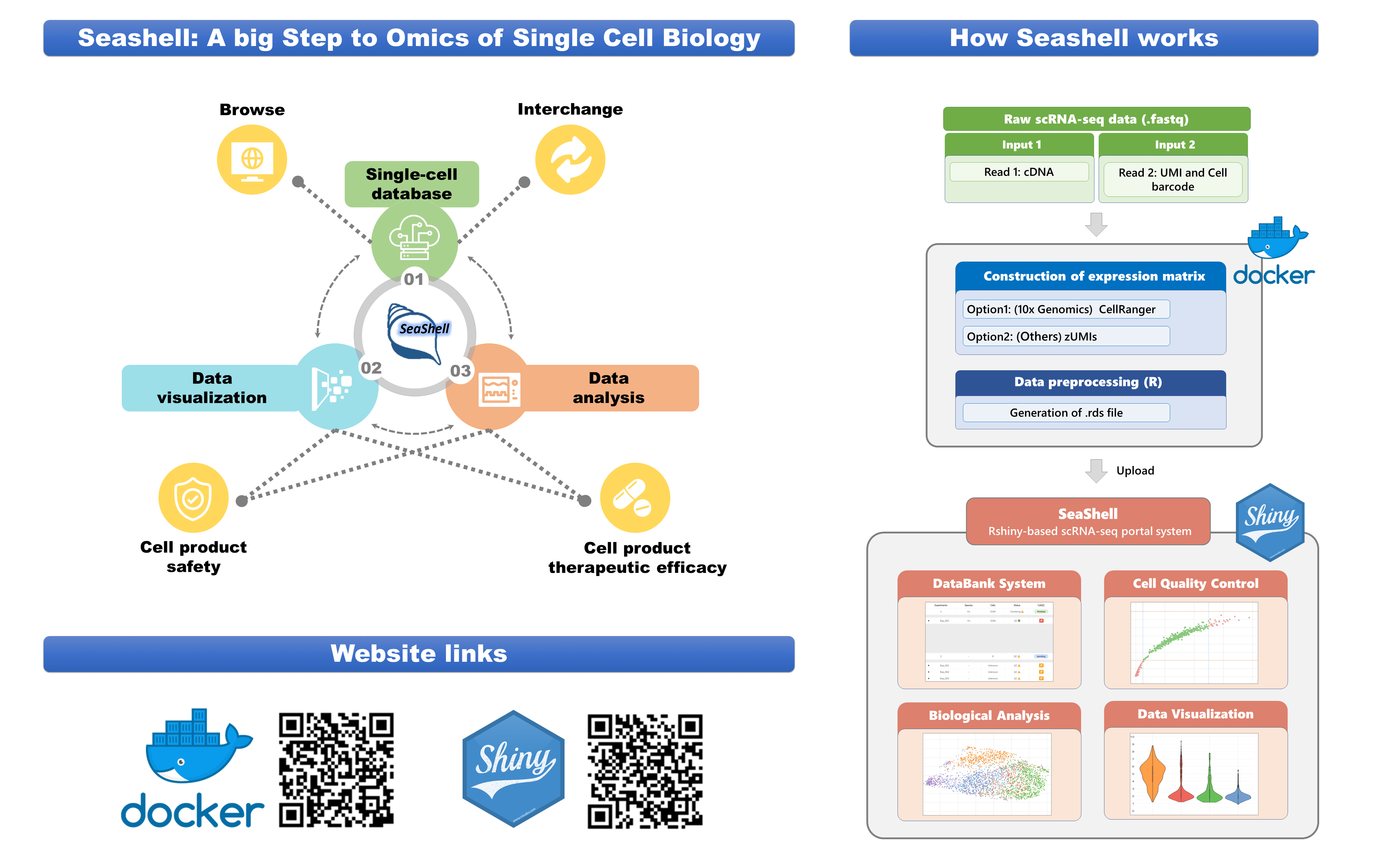
| Scientific Breakthrough | The characteristics of scRNA-seq analysis platform Seashell contain : (1) the custom pipeline of data analysis from raw data (.fastq) (2) the user-friendly interface of a single-cell database. (3) the integrated visualization platform which contains various tools. |
||
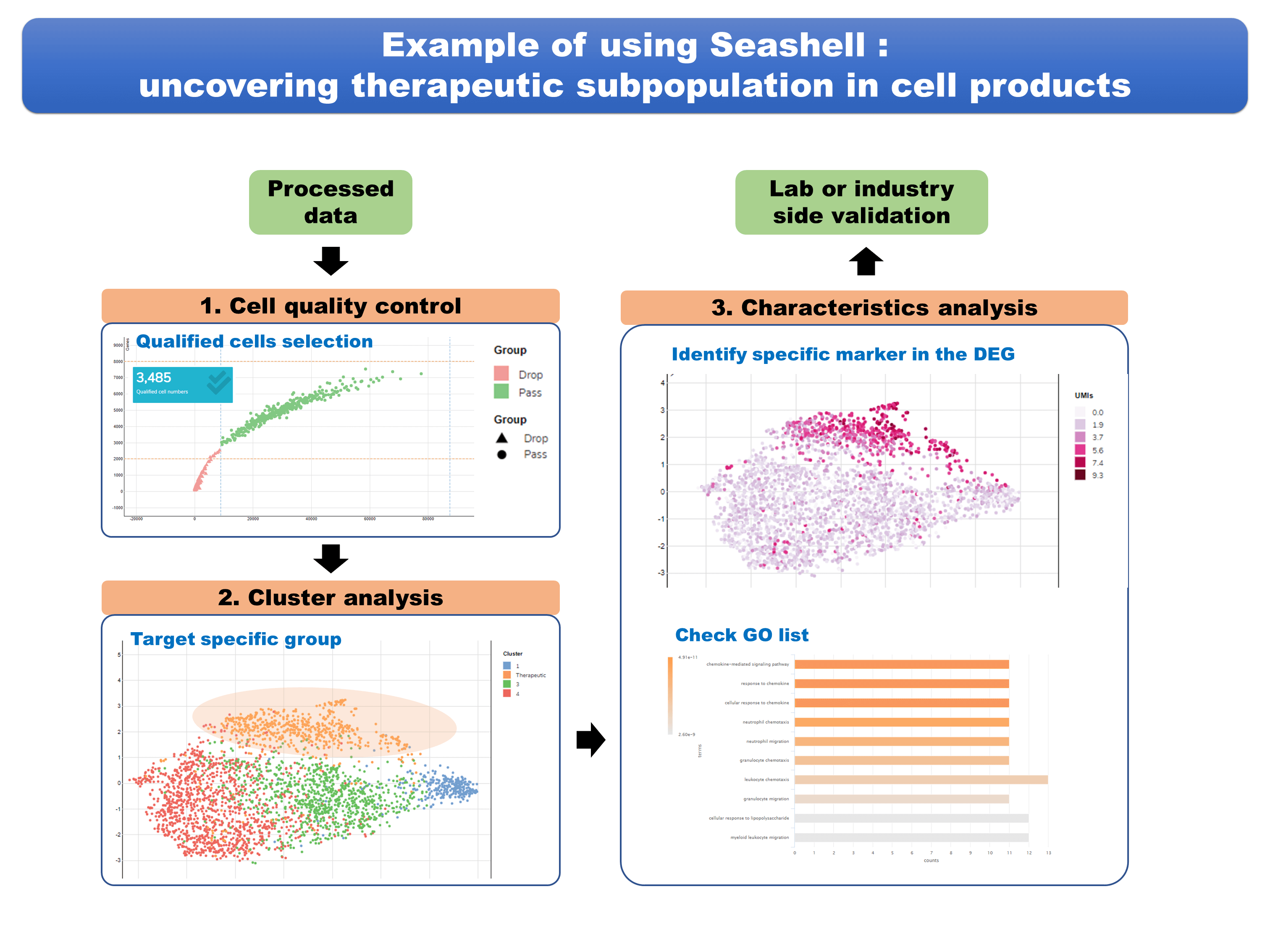
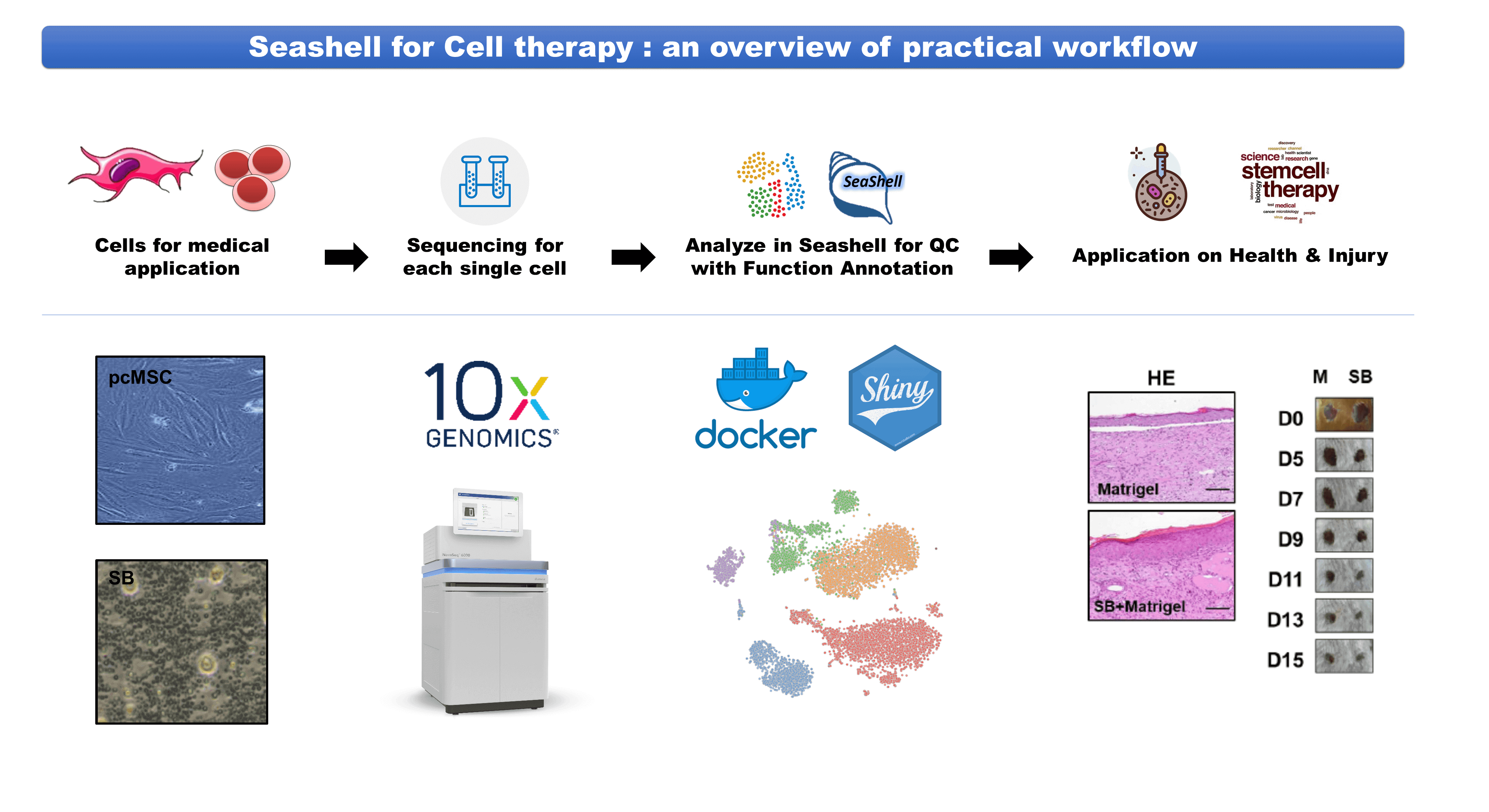
| Industrial Applicability | Our research group cooperated with the cell therapy groups in the Taipei Medical University and National Taiwan University College of Medicine, performing data analysis for high dimensional single-cell transcriptomics data of two potential cell products by our Seashell platform. We successfully obtained the safety and therapeutic information of these products and demonstrated the performance of the web portal system. We also plan to apply this web-based platform to various research and industrial fields in the future. |
||
| Keyword | single-cell RNA sequencing transcriptome docker galaxy droplet-seq cell therapy web application biomarker identification precision medicine big data | ||
- cylin@iis.sinica.edu.tw
other people also saw