| Technical Name | Cell-free virus-host chimera DNA from hepatitis B virus integration sites as a circulating biomarker for monitoring hepatocellular carcinoma | ||
|---|---|---|---|
| Project Operator | National Taiwan University Centers of GenomicPrecision Medicine | ||
| Project Host | 陳培哲 | ||
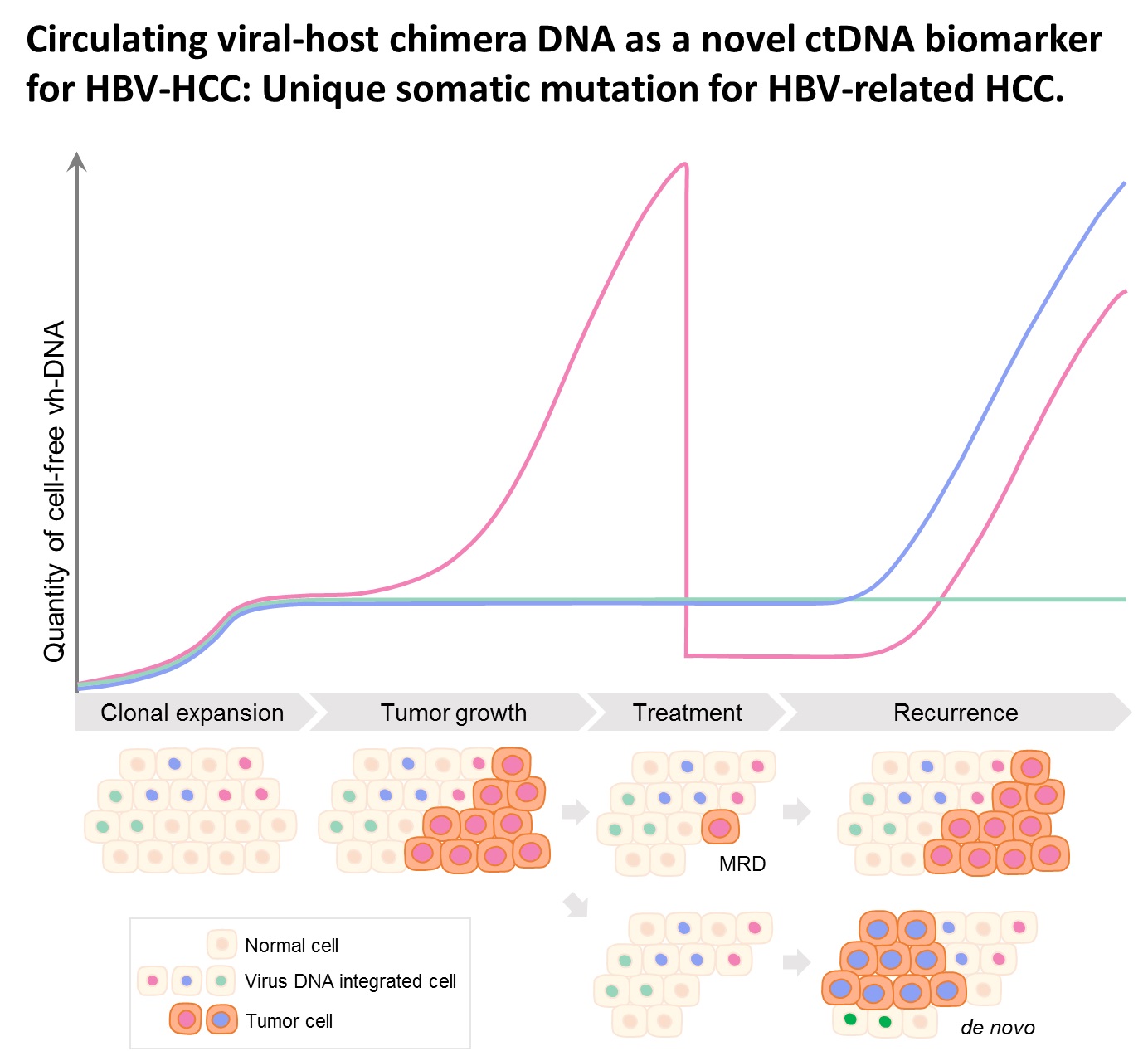
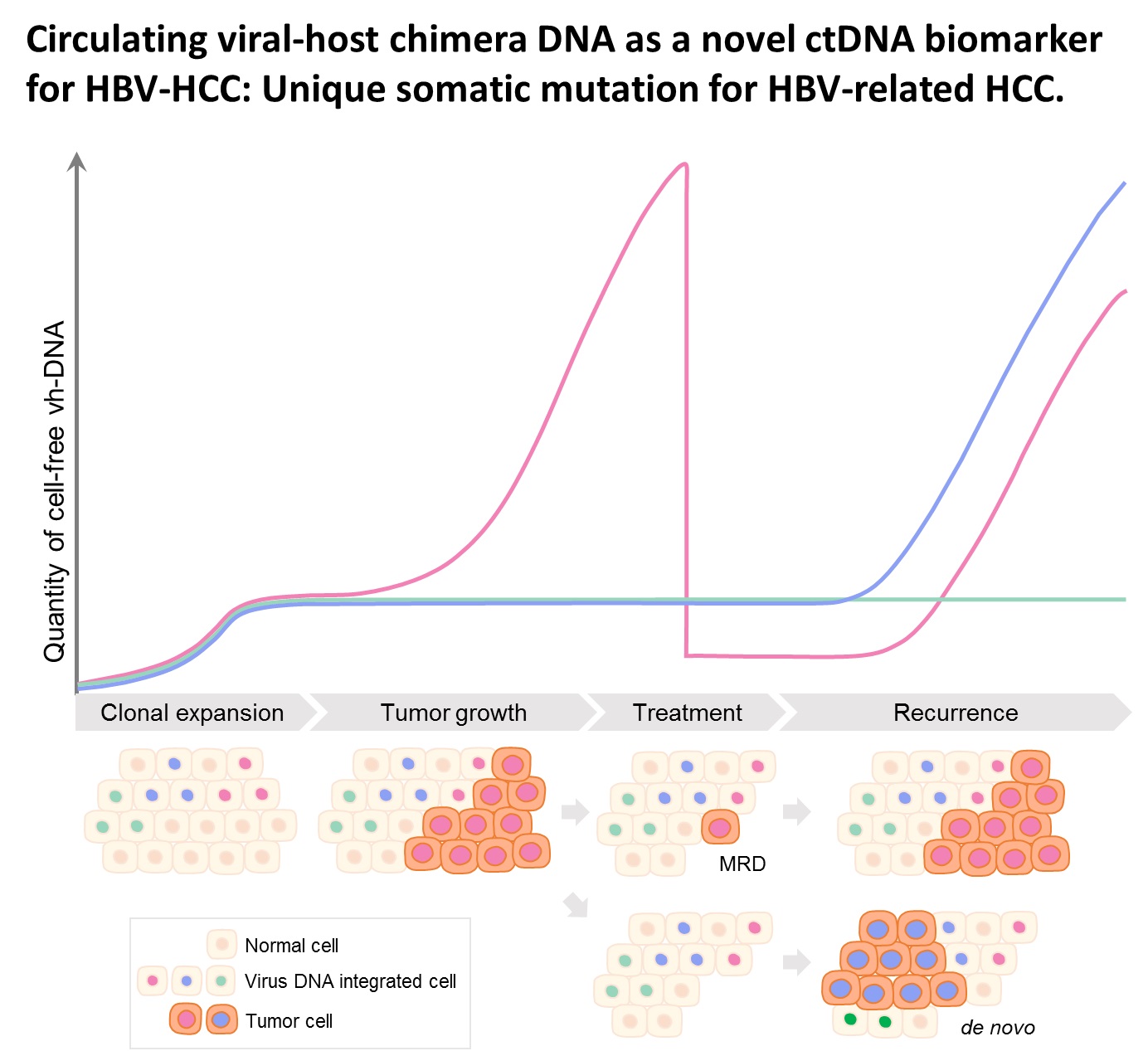
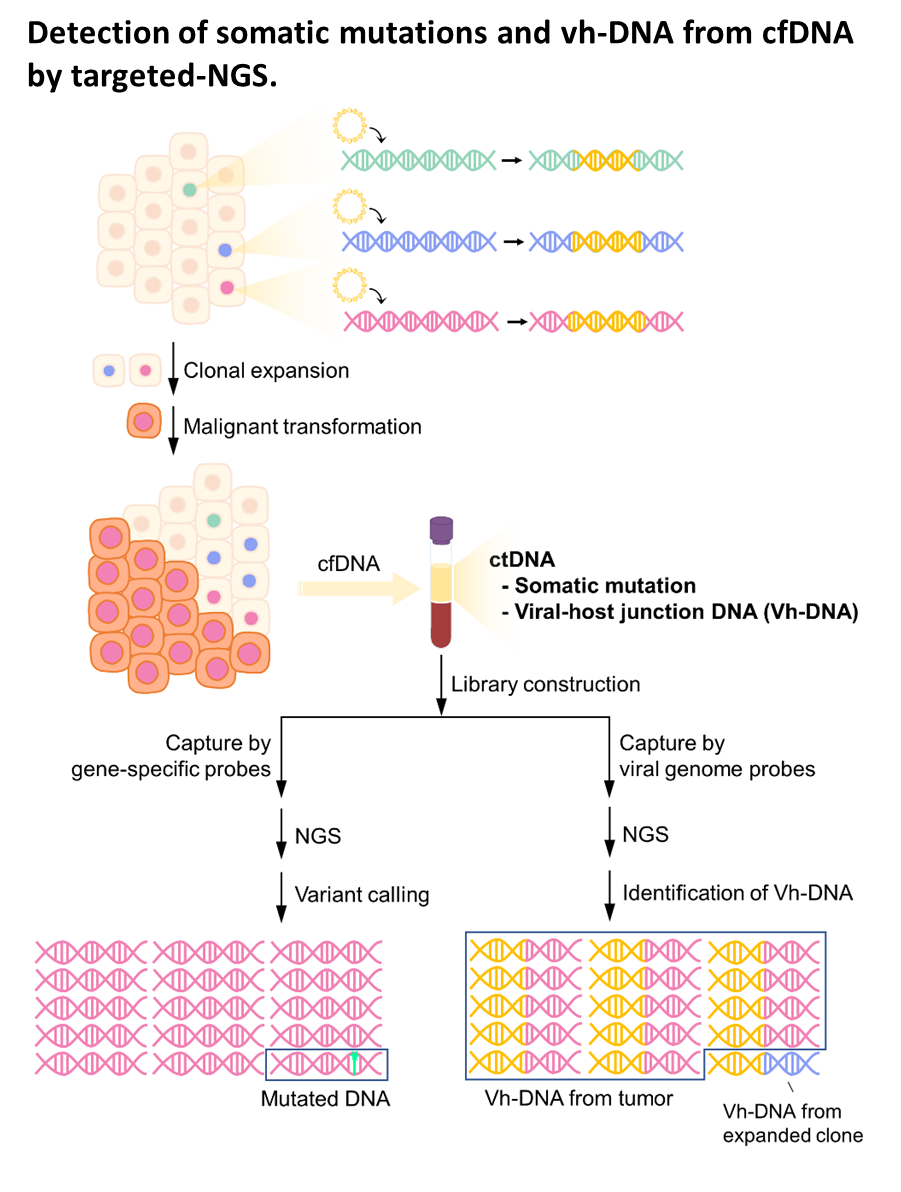
| Summary | HBV integration in patients with HBV-related HCC was determined by the hybridization capture-based next-generation sequencing (Capture-NGS) platform, with DNA from plasma samplesHCC tissues. For individual HCC, the vh-DNA was validatedquantified by specific droplet digital PCR (ddPCR) assay, which is developed to measure the corresponding vh-DNA for follow up of the recurrence of HCCeven the development of primary HCC. |
||
| Scientific Breakthrough | The vh-DNA provides a more sensitive way for monitoring HCC than the protein type of biomarker, AFPPIVKA-II, with detection rate of 90 versus 30-40 for tumor 3-5 cm. In addition, the vh-DNA overcomes the limitations in using somatic point mutations as cell-free tumor markers. First, the detection of vh-DNA is not competed by a high level of wild-type background in total cfDNA released from normal tissues. Second, the vh-DNA covers 90 of HBV-HCC, much higher than the common point mutations (30-50). Third, vh-DNA provides a unique barcode for individual tumors for determination of the clonal origin of recurrence. Fourth, detection of vh-DNA in in tumor screening of healthy subjects indicates the HCC regarding to virus tropism. |
||
| Industrial Applicability | This platform technology is already processed for technology transferundergoing clinical trial, which will be completed in near future. We expect the results will help validate its clinical application for the detection of recurrent HCC after tumor resection. |
||
| Keyword | Cell-free DNA hepatocellular carcinoma hepatitis B virus virus-host chimera DNA recurrent HCC | ||
- Contact
- Yee-Shin Lee
- yslee@ntu.edu.tw
other people also saw