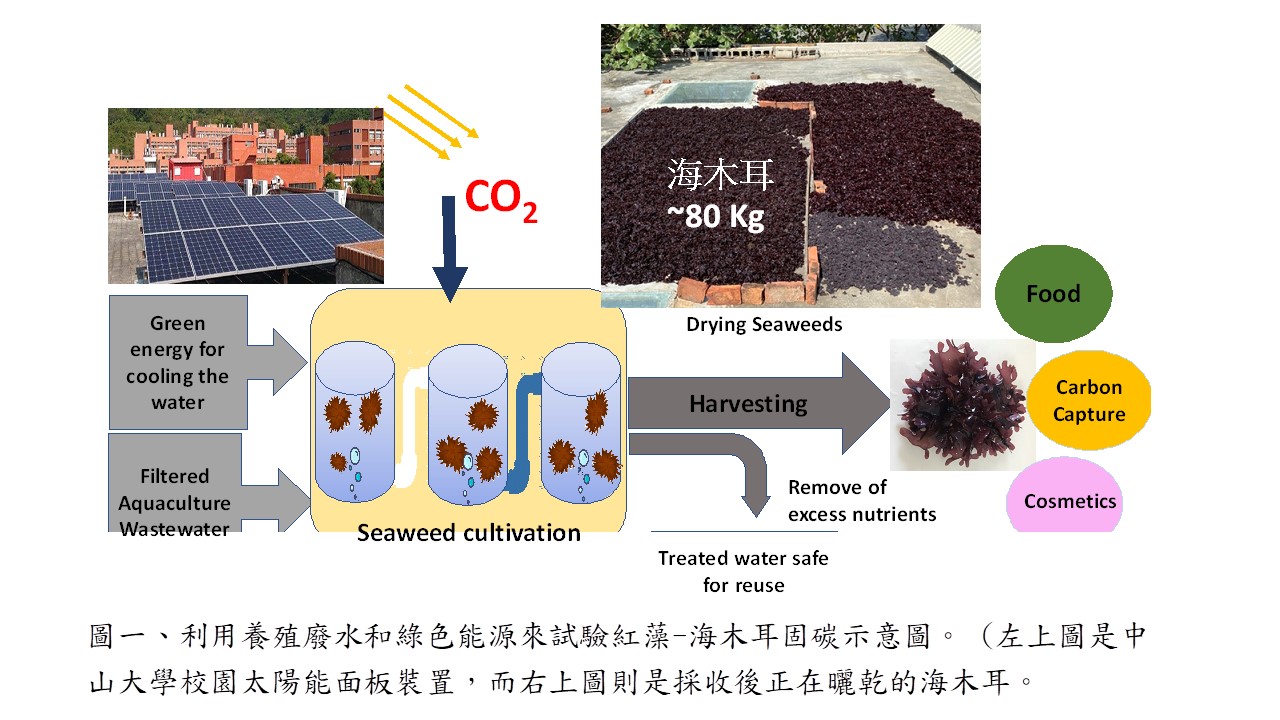
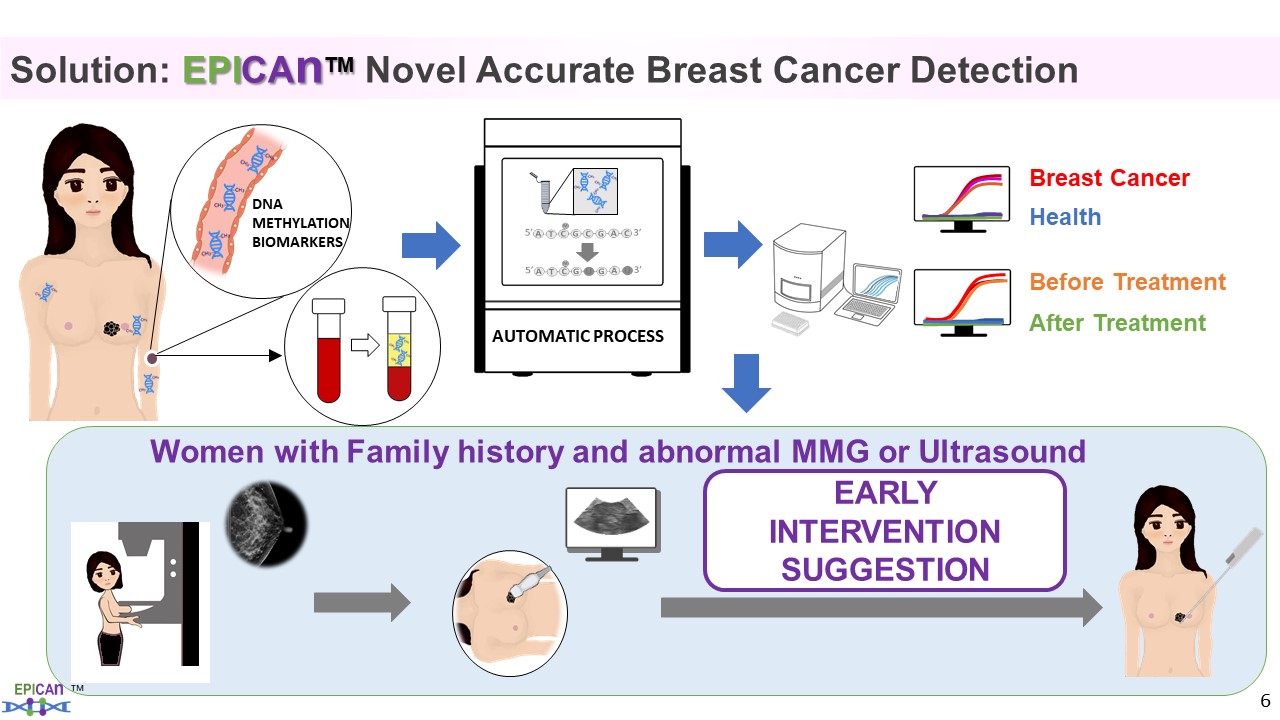
| Summary |
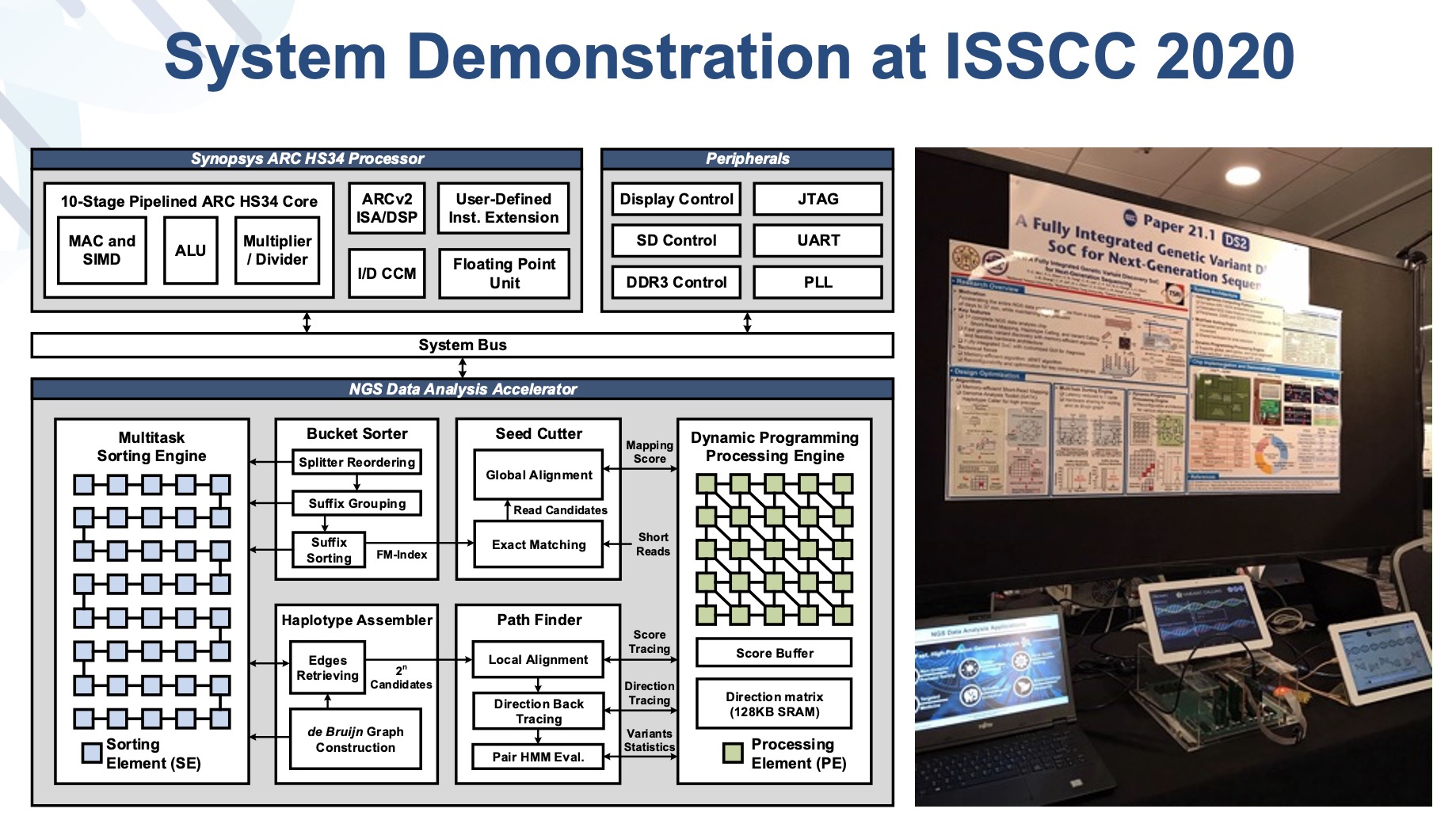
This research presents a genetic variant discovery SoC for analyzing Next-generation sequencing data which is embedded with the processorconnected with some peripherals. With hardware-optimized algorithm sBWT, we greatly speed up the process of short reads mapping by k-ordered FM-indexsuffix grouping. After short reads mapping, we utilized de bruijn graph for sequence assembly to rebuild haplotypes,built haplotypes are compared to the reference genome to detect genome variations. The detected variations are written to files for further clinical diagnosisresearch analysis.
|
| Scientific Breakthrough |
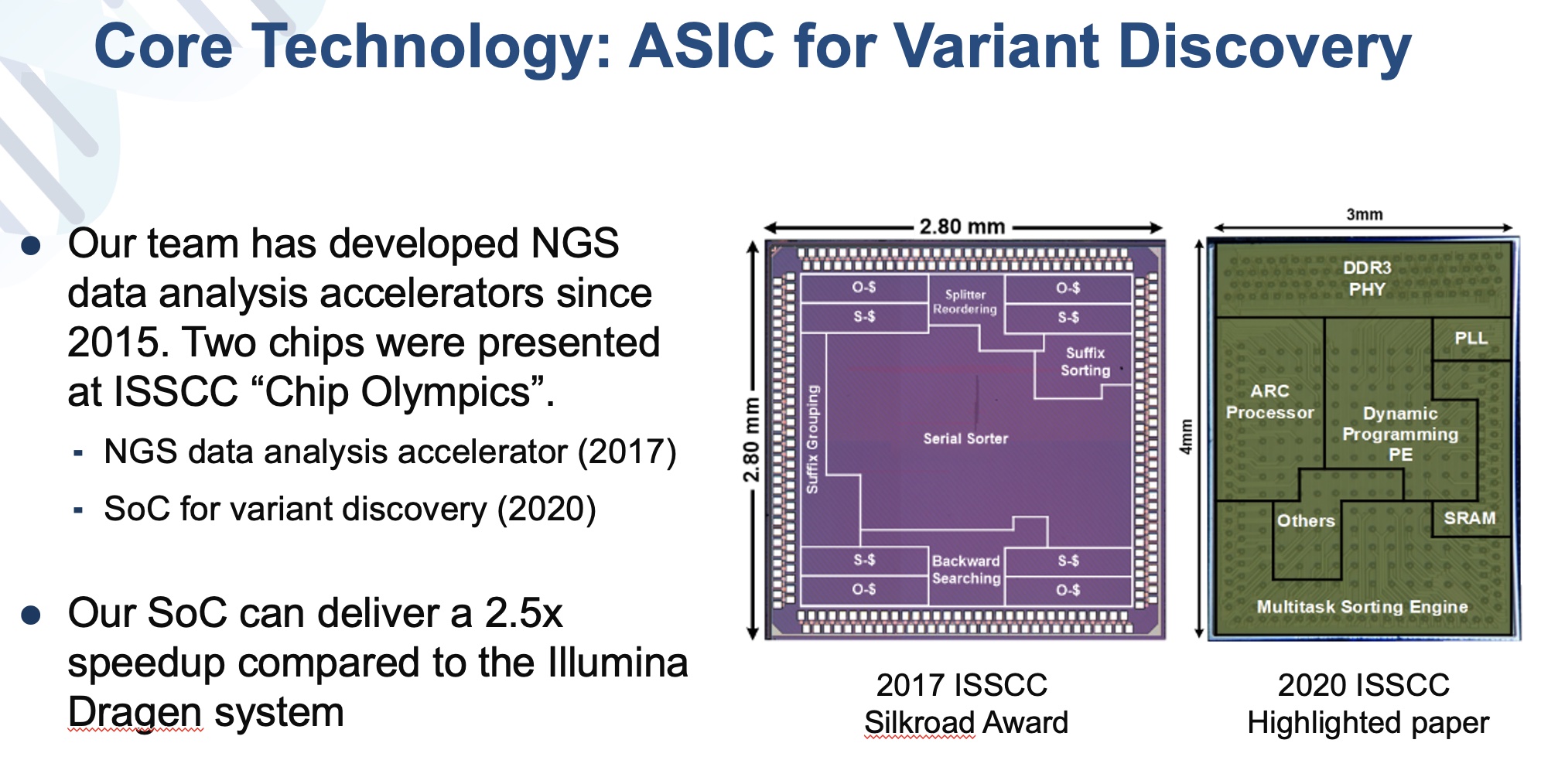
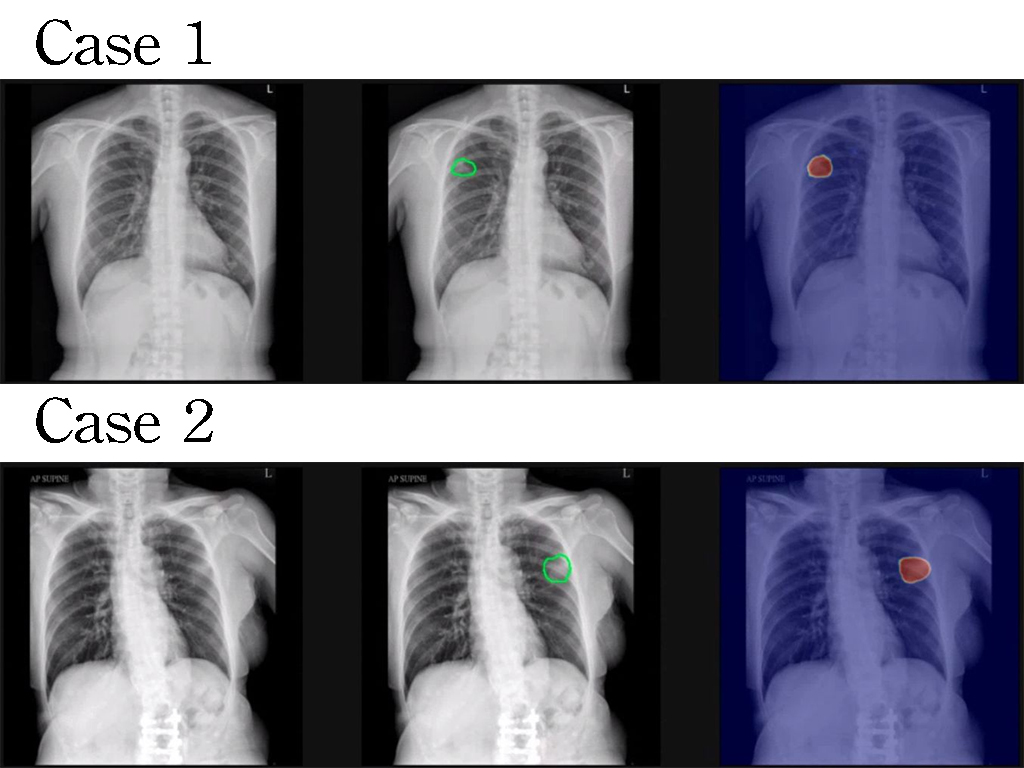
Existing NGS data analysis circuits mostly focus on optimizing part of the analysis pipeline. Our work integrates the entire data analysis pipeline,becomes the first completefast NGS data analysis platform. Comparing to existing high-level GPU platform, our work achieved 66 times speed-up,had 150003000 gains on energy efficiencyarea efficiency, correspondingly. Our work achieved an accuracy of 99.6 when evaluated on test data provided by FDA.
|
| Industrial Applicability |
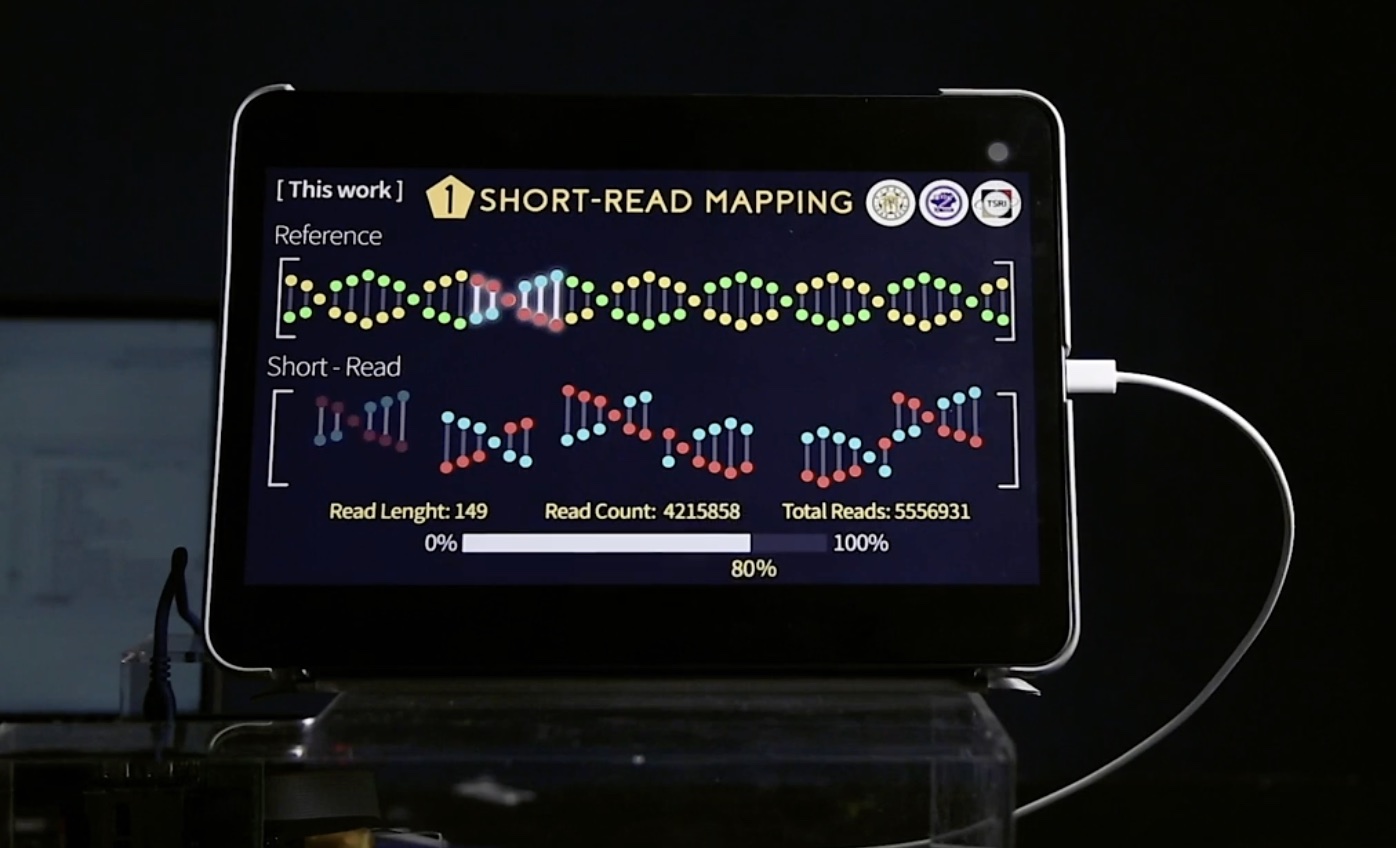
The market value of next-generation sequencing industry is by acclamation to have exponential growth. However, downstream analysis for sequencing data is time-consuming, taking several dayseven weeks which is not capable of in-time clinical diagnoses. To deal with this issue, our team developed Soc which is especially for variant calling,greatly accelerated sequencing data analysis. By using our Soc, the entire analysis can be done in no more than 40 minutes. In addition, our Soc is fully portable, hence is able to be brought to clinical situation for in-time clinical diagnoses.
|